
Next: Data
Up: Data Types
Previous: Data Types
type unit_cell
real(kind=dp), dimension(3,3) :: real_lattice
real(kind=dp), dimension(3,3) :: recip_lattice
real(kind=dp) :: volume
real(kind=dp), dimension(:,:,:), pointer :: ionic_positions
real(kind=dp), dimension(:,:,:), pointer :: ionic_velocities
logical, dimension(:,:,:), pointer :: atom_move
integer :: num_ions
integer :: num_species
integer :: max_ions_in_species
integer, dimension(:), pointer :: num_ions_in_species
integer, dimension(:), pointer :: ionic_charge
real(kind=dp), dimension(:), pointer :: species_mass
character(len=10), dimension(:), pointer :: species_symbol
character(len=file_maxpath), dimension(:), pointer :: species_pot
integer, dimension(:), pointer :: species_lcao_states
integer, dimension(:), pointer :: ion_pack_species
integer, dimension(:), pointer :: ion_pack_index
read(kind=dp), dimension(:,:), pointer :: initial_magnetic_moment
end type
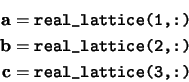
real_lattice The real space lattice vectors defining the unit cell. The real space cell lattice vectors are stored in rows of the array
real_lattice. I.e.
recip_lattice The reciprocal space lattice vectors defining
the Brillouin zone. The reciprocal space lattice vectors are stored in
the rows in the same way as the real space lattice vectors.
volume The volume of the unit cell.
ionic_positions The positions of the ions in the unit
cell. The positions of the ions in the cell are stored in fractional
coordinates in the basis of the real space lattice vectors. For any
species ns, only the elements
ionic_positions(:,1:num_ions_in_species(ns),ns) are valid.
ionic_velocities The velocities of the ions in the unit
cell. The velocities of the ions in the cell are stored in Cartesian
coordinates. For any species ns, only the elements
ionic_velocities(:,1:num_ions_in_species(ns),ns) are valid.
atom_move Logical flag to indicate if the coordinate of
a each ion is fixed by the ionic constraints.
num_ions The total number of ions in the unit cell.
num_species The number of species in the unit cell.
max_ions_in_species The maximum number of ions in any one species.
num_ions_in_species The number of ions in each species.
ionic_charge The charge of the bare ion of each species.
species_mass The mass of each species.
species_symbol The symbol representing each species.
species_pot The filename of the pseudopotential representing
each species.
species_lcao_staes The number of angular momentum states to use in the LCAO basis set for this species for the purpose of population analysis.
ion_pack_species This array defines the ordering of the ionic species if data are held in a `packed' array, i.e. a one-dimensional array of length num_ions. An ion at position 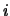 is of species
is of species ion_pack_species(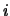 ).
).
ion_pack_index This array defines the ordering of the ions within a species if data are held in a `packed' array, i.e. a one-dimensional array of length num_ions. An ion at position 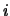 is index
is index ion_pack_index(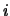 ) within its species.
) within its species.
initial_magnetic_moment This array stores the atomic
magnetic moments, set with the MAGMOM flag in the POSITIONS_* block of
the cell file and used to initialise the spin density of the system.
Conditions

-
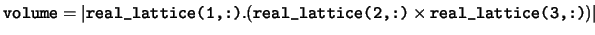

-
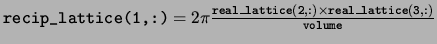

-
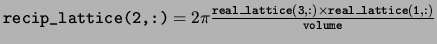

-
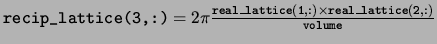

-
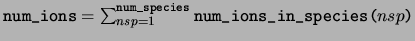

-
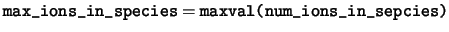

-
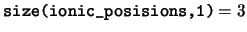

-
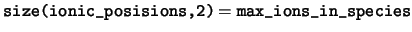

-
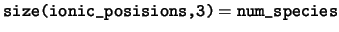

-
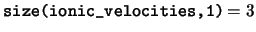

-
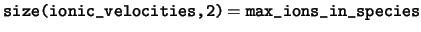

-


-
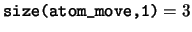

-
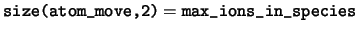

-
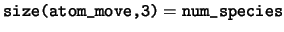

-
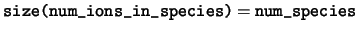

-
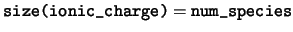

-
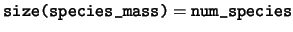

-
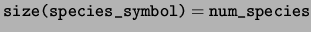

-
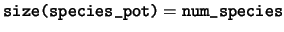

-


-
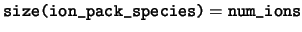

-
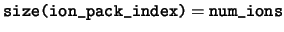

-
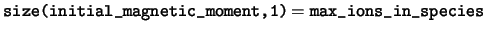

-
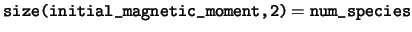
The unit_cell data type defines the dimensions and contents
of the unit cell.
Maintaining the consistency of the real and reciprocal lattice vectors
and cell volume is the responsibility of the programmer using the
module if a cell is changed. Subroutines are provided to aid in
this.
The order in which the data for the species occur in
ionic_positions, num_ions_in_species,
ionic_charge, species_mass, species_symbol,
species_pot and species_lcao_basis must be the same.


Next: Data
Up: Data Types
Previous: Data Types
Jonathan Yates
2004-04-16